Evolution and Information:
The Nylon Bug
![]()
Evolution and Information:
The Nylon Bug
![]()
by Dave Thomas : nmsrdaveATswcp.com (help me fight SPAM! Please replace the AT with an @ )
Creationists often say that all mutations are harmful and deleterious, and degrade the genome. They say that mutations can only scramble the information that's there, and that mutations cannot produce new "information." This page shows why they are wrong.
![]()
Wondering about Answers in Genesis and their "refutation" of the Nylon Bug as due to plasmids? - Click here for an update!
![]()
How about Answers in Genesis and the TJ Vol. 17 #3 article "The adaptation of bacteria to feeding on nylon waste" by Don Batten? Click here for an update!
![]()
Lee Spetner, author of Not By Chance, has taken on the Nylon Bug. Click here for an update!
![]()
The Nylon Bug
My favorite example of a mutation producing new information involves a Japanese bacterium that suffered a frame shift mutation that just happened to allow it to metabolize nylon waste. The new enzymes are very inefficient (having only 2% of the efficiency of the regular enzymes), but do afford the bacteria a whole new ecological niche. They don't work at all on the bacterium's original food - carbohydrates. And this type of mutation has even happened more than once!
So, what is a frame shift mutation?
It happens when a chunk of genetic code (remember those AGTCTAGATCGTATAGC... DNA sequences from Jurassic Park?) is shifted by one or more nucleotides. In DNA, each triplet of nucleotides codes for one amino acid, and each such triplet is called a codon. So, the amino acid Arginine (symbol Arg) is coded by the DNA nucleotide sequence CGT, and also by codons CGA, CGC,CGG, AGA, AGG. Likewise, the amino acid Glutamic Acid (symbol Glu) is coded by the DNA nucleotide sequence GAA, and also by the sequence GAG. There are four types of nucleic acids, which naturally bond in one of two pairs: Thymine/Adenine, and Cytosine/Guanine (T/A and G/C). A thymine (T) on one strand of DNA will bind to an adenine (A) on the paired strand, and so on. There would be 64 different possible amino acids with a three-nucleotide codon (43=64), but several of these are redundant, as shown in the lists above for amino acids Arginine and Glutamic Acid. In biological organisms, there are just 20 different amino acids. Various DNA triplets code for these amino acids, and strings of amino acids form proteins - molecules (such as enzymes) that really do something specific, such as metabolize sugars.
A Frame Shift is a radical mutation in which a single nucleotide is inserted or deleted, causing a shift in the triplets coded by the DNA strand. It's fairly technical, so I'll present what a Frame Shift is by analogy with a different Digital Code, that being the ASCII code used in computers to convert numbers from 0 to 255 into symbols or characters. For example, the ASCII code for the letter "A" is 65, which in binary converts to 64+1, or 26 + 1, written thus: 01000000 + 00000001 = 01000001. For this analogy, we'll just be using the first 128 characters, and so we can use just 7 digits: thus, an "A" then has the 7-digit code 1000001. A lower case "a" is 32 higher than a capital A (which leaves room for 26 letters and a few extra characters), and is thus written 1100001 in 7-digit binary notation (=64+32+1 = 97 in decimal). A "b" is written 1100010 in 7-digit binary notation (=64+32+2 = 98). Likewise, a "d" is written 1100100 in 7-digit binary notation (=64+32+4 = 100), and an "e" is written 1100101 in 7-digit binary notation (=64+32+4+1 = 101).
What has all this to do with Frame Shifts, you ask? In this analogy, actual biological proteins or enzymes (strings of amino acids) correspond to words or phrases (strings of ASCII characters). Individual amino acids (such as Arginine) are analagous to individual ASCII characters (such as the letter "A"). Finally, the DNA nucleotides A, T, C and G correspond to the binary digits 0 and 1.
So, let us string together several letters to make a "digital" word. The ASCII digital code for the word "bed" is made by stringing together the 7-digit codes for b (1100010), e (1100101), and d (1100100) to make one long code: 110001011001011100100.
The image below shows what happens when we apply a Frame Shift to the digital code for bed. Here, we shift the "reading frame" by one digit to the left, which requires that we add one extra digit as a prefix. Here, the prefix I chose was the digit 1.
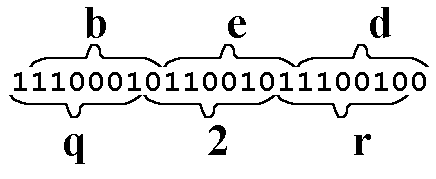
The Frame Shift is not a mild mutation. It is HUGE. We still have a 3-letter string, but each letter is different. Shifting the reading frame one digit gives us three NEW characters: q:(1110001), 2 (0110010), and r (1110010).
This particular Frame Shift scrambles the perfectly fine word "bed" into the unintelligible, meaningless word "q2r." In this case, the Frame Shift is not only a drastic mutation, but has completely altered the meaning of the word "bed." In this case, at least, information has been "lost"or "degraded," just as creationists say will happen ALL THE TIME - EVERY TIME.
And that's where they are wrong. While most Frame-Shift mutations do indeed scramble meanings and degrade information, not all of them do so.
Here's an example of a frame shift creating information: here, the word "gas" is coded as g(1100111) + a (1100001) + s (1110011). When we apply a Left Frame Shift to the long code for "gas," we do NOT end up with a meaningless phrase such as "q2r." In THIS case, we end up with a new, meaningful word: spy.
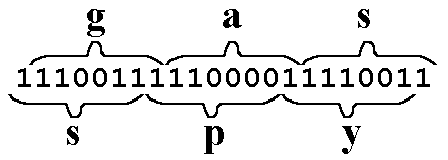
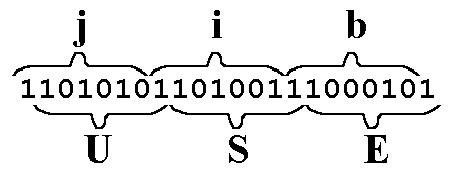
Similarly, the word "jib," when right-frame-shifted, is mutated into the new word "USE."
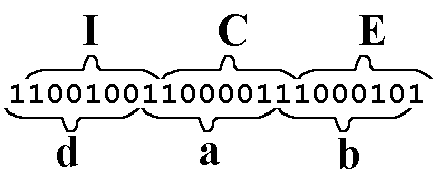
As a final example, the word "ICE," when left-frame-shifted, is mutated into the new word "dab."
Certainly, MOST frame shifts will destroy information. BUT NOT ALL - and that is where creationists have it wrong. I have shown three examples where such "Frame Shifts" indeed create new information. After all, in the proper context, the words "spy," "USE," and "dab" actually mean something. Since their meanings are totally unrelated to the original meanings, it is obvious that, at least in this case, the Frame Shift mutation process has created new information. It's important to note that context really means something as regards interpretation of these words. For example, if the word "luz" was generated, that would mean nothing in English, but it means "light" in Spanish. Without a common language and culture, words won't mean anything! It's different with DNA, because the "context" in which DNA strands are interpreted is the world of chemical reactions. The "meaning" of novel strands of DNA lies in how these strands are transcribed, what the new proteins look like, and (most importantly!) how the proteins react with other molecules, perhaps even affecting the organism's lifestyle.
Now, let's get back to Biology, and the case of the bacterium which has evolved the capability of ingesting nylon waste (see Kinoshita et. al.). This case is most interesting. Nylon didn't exist before 1935, and neither did this organism. Detailed examination of the DNA sequences of the original bacterium and of the nylon-ingesting version show identical versions in the gene for a key metabolic enzyme, with only one difference in over 400 nucleotides. However, this single microevolutionary addition of a single thymine ('T') nucleotide caused the new bacterium's enzyme to be composed of a completely novel sequence of amino acids, via the mechanism of frame shifting. The new enzyme is 50 times less efficient than its precursor, as would be expected for a new structure which has not had time to be polished by natural selection. However, this inefficiency would certainly not be expected in the work of an intelligent designer. The genetic mutation that produced this particular irreducibly-complex enzyme probably occurred countless times in the past, and probably was always lethal, until the environment changed, and nylon was introduced.
The image below shows just a part of the 400+-long nucleotide string for the key enzyme (see the Susumu Ohno paper). The original ("old") enzyme's amino acid sequence appears on top, and the frame-shifted ("new") sequence on bottom. The DNA nucleotides appear in the middle for both the old species and the new (one T inserted). Over this small portion of the enzyme, the old DNA coded for the amino acids Arginine, Glutamic Acid, Arginine, Threonine, Phenylalanine, Histidine, Arginine and Proline.

But the NEW DNA strand, which includes one extra T nucleotide, is shifted, and the new string of amino acids is completely changed. The addition of the thymine nucleotide produces a new Methionine amino acid, which, like the conductor tapping his baton, indicates the Start of a new Protein. This is followed by other new amino acids because of the frame shift: Asparagine, Alanine, Arginine, Serine, Threonine, Glycine and Glutamine. The new string of amino acids - the new protein - is completely different from the original.
While most frame shifts of such a key enzyme would destroy the enzyme, resulting in immediate death of the organism, this particular protein happened to react with nylon oligomers. And so it was that a drastic mutation suddenly gave an ordinary sugar-eating bacterium the unusual ability to digest nylon, which just happened to be present in abundance in the little waste pond behind a Japanese factory. The Japanese scientists who discovered strange bacterial mats growing in their scum ponds became very interested in this new ability, and finally found it was all due to a single Frame Shift mutation. The new enzyme is not active on common substrates - the bacteria's old "food" - and plenty were checked. Whether or not these bacteria retain enzymes to digest their former food source, the fact is that the former food source became much less important because of the new-found ability to ingest food from a novel source - nylon waste.
The creationist argument that all mutations must destroy information is clearly wrong. In this case, a mutation has clearly produced new information. That is, unless you want to quibble that the detailed three-dimensional structure and composition of a protein that reacts specifically to nylon is not "information."
Creationists usually counter this example by claiming that the bacterium is, after all, "still a bacterium." It didn't mutate into a whale or a dinosaur. But that's changing the subject. The subject of this essay is "Can Mutations Create New Information."
Science and logic both show the answer is a resounding YES.
Key points to ponder:
References
"Purification and Characterization of 6-Aminohexanoic-Acid-Oligomer Hydrolase of Flavobacterium sp. K172," Kinoshita, et. al., Eur. J. Biochem. 116, 547-551 (1981), FEBS 1981.
Key Quote: "There are two possible reasons for an enzyme to be active on an unnatural substrate: one is that an unnatural compound could be decomposed by an enzyme if it were an analogue of that enzyme's physiological substrate, and the other is that an unnatural substrate could be decomposed by a newly evolved enzyme. The data obtained in this study show that 6-aminohexanoic-acid-oligomer hydrolase has no activity on any physiological substrates, including the linear and cyclic amides and peptides tested..."
"Birth of a unique enzyme from an alternative
reading frame of the pre-existed, internally repetitious coding
sequence", Susumu Ohno, Proc. Natl. Acad. Sci.
USA, Vol. 81, pp. 2421-2425, April 1984.
OHNO ON-LINE: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=345072
COMPLETE PDF: http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=345072&action=stream&blobtype=pdf
"New Proteins without God's Help." Thwaites, W., Creation/Evolution 1985; 5(2), Issue 16: 1-3.
UPDATE: Answers In Genesis Dismisses the Nylon Bug as simply due to "Plasmids."
From http://www.answersingenesis.org/home/area/feedback/negative7-24-2000.asp :
"... Finally, Mr Cerutti is out of date about this new nylon digesting ability allegedly from a frame shift. New evidence shows that the ability was due to plasmids [e.g. K. Kato, et al., ‘A plasmid encoding enzymes for nylon oligomer degradation: Nucleotide sequence analysis of pOAD2’, Microbiology (Reading) 141(10):2585–2590, 1995. In fact, more than one species of bacteria have the ability, residing on plasmids. This suggests that the information probably already existed, and was just passed between different types of bacteria. ..."
Sorry, AiG, but just because something is on a plasmid doesn't mean it's always been there! In fact, the plasmid involved in this case is very well known and characterized. Scientists have studied both the original (pre-mutation) plasmid and the novel (post-mutation) plasmid, in great detail. It turns out that the novel plasmid's mutated DNA for production of nylonase is almost identical to a non-coding repetitive DNA sequence on the original plasmid; the difference is the single nucleotide that triggered the Frame Shift. This mutation did not exist 60 years ago. If this gene was always there, whether in a plasmid or not, we can reasonably wonder why a bacteria would have a gene for hydrolysing an artificial polymer that did not exist until just a few decades ago; and why, in the absence of such a substrate, was the gene not mutated to uselessness over the millenia?
Was the plasmid slipped in from another bacterium? NO!! The plasmid in question, pOAD2, is just one of three plasmids that are harbored by the bacterium under investigation here, Flavobacterium Sp. K172. Here are some citations to back this up:
"Sequence analysis of a cryptic plasmid from Flavobacterium sp. KP1, a psychrophilic bacterium," Makoto Ashiuchi, Mia Md. Zakaria, Yuriko Sakaguchi, Toshiharu Yagi, FEMS (Federation of European Microbiological Societies) Microbiology Letters 170 (1999), 243-249.
"Bacteria of genus Flavobacterium, Gram-negative bacteria, are widely distributed in soil and fresh marine waters. Some of them harbor plasmid(s) involved in metabolism of synthetic organic compounds. Flavobacterium sp. K172 harbors plasmids, pOAD1, pOAD2 and pOAD3; pOAD2 (43.6 kbp) encodes nylon oligomer degradation genes."
"A New Nylon Oligomer Degradation Gene (nylC) on Plasmid pOAD2 from a Flavobacterium sp.," Seiji Negoro, Shinji Kakudo, Itaru Urabe, and Hirosuke Okadam, Journal of Bacteriology, Dec. 1992, p. 7948-7953.
"The EI-encoding gene (F-nylA) and EII-encoding gene (F-nylB) of Flavobacterium sp. K172 are located on plasmid pOAD2 (44 kb), one of the three plasmids harbored in strain K172."
"Birth of a unique enzyme from an alternative reading frame of the pre-existed, internally repetitious coding sequence", Susumu Ohno, Proc. Natl. Acad. Sci. USA, Vol. 81, pp. 2421-2425, April 1984. PDF
"Analysis of the published base sequence residing in the pOAD2 plasmid of Flavobacterium sp. K172 indicated that the 392-amino acid-residue-long bacterial enzyme 6-aminohexanoic acid linear oligomer hydrolase involved in degradation of nylon oligomers is specified by an alternative open reading frame of the preexisted coding sequence that originally specified a 472-residue-long arginine-rich protein."
It's interesting to note that the precise plasmid of Flavobacterium sp. K172, namely pOAD2, was cited by Susumu Ohno fully eleven years before the publication of the "new evidence" that AiG claims " shows that the ability was due to plasmids..."
The Bottom Line: Just because this mutation wasn't confined to a cell's main chromosomes does not mean it didn't happen. (A plasmid is defined as a replicon - a replicating piece of DNA - that is inherited in an extrachromosomal state.) This case still provides an excellent example of a New Protein that evolved without the assistance of an Intelligent Designer.
(My thanks to Ian Musgrave and Ian Ferguson for useful discussions on this topic).
UPDATE: Don Batten of "Answers In Genesis" writes on "The adaptation of bacteria to feeding on nylon waste" in the Technical Journal, Vol. 17, Issue 3 (December 2003)
From http://www.answersingenesis.org/tj/v17/i3/bacteria.asp?vPrint=1
"However, there are good reasons to doubt the claim that this is an example of random mutations and natural selection generating new enzymes, quite aside from the extreme improbability of such coming about by chance. ..."
Batten's article is dismantled a plank at a time by Ian Musgrave in the Talk.Origins Post of the Month for April 2004, "Nylonase Enzymes", http://www.talkorigins.org/origins/postmonth/apr04.html .
Musgrave's bottom line:
"Generation of the nylon hydrolysing genes is standard 'mutation followed by selection.' The AiG article shows once again how poor their understanding of both biology and evolutionary theory is. "
UPDATE: Lee Spetner, author of "Not By Chance" comments on "TheNylon Bug" on Arthur S. Lodge's website (November 2002)
"It's interesting, first of all, that the URL you pointed to picked the "nylon bug" as an example of a random mutation yielding a gain of information. (The short answer is, the mutation does yield an increase of information, but was it random?) . ... Let me point out two important facts that the URL [www.nmsr.org/nylon.htm] ignores. First, there are two altered enzymes, not just one. Both these enzymes are needed to metabolize the 6-aminohexanoic-acid-cyclic-dimer (6-AHA CD) found in the waste water of the nylon factory. Neither of these enzymes alone is effective. Both are needed. The first enzyme, which I shall call enzyme 1, is 6-aminohexanoic-acid-cyclic-dimer hydrolase (6-AHA CDH) and catalyzes the conversion of 6-AHA CD to 6-aminohexanoic-acid-oligomer (6-AHA LO). The second enzyme, which I shall call enzyme 2, is (6-aminohexanoic-acid-oligomer hydrolase (6-AHA LOH) and catalyzes the conversion of 6-AHA LO to 6-amino-hexanoic acid [Kinoshita et al. 1981]. Only enzyme 2 is the product of a frame shift. Enzyme 1, whose DNA sequence I have not seen, is probably the product of only point mutations. [Okada et al. 1983, Ohno 1984]Second, enzyme 2 is not just the product of a frame shift, it is also the product of 140 point mutations. Many of these mutations are silent, but many are not. 47 amino acids out of 392 of the enzyme have been changed.
It seems to me that many of these altered amino acids are essential to the catalytic effect of the enzyme. How many, I don't know. In my above cited letter to Jim, I calculated the probability of getting multiple random mutations in the 30 years it took to evolve these enzymes. If the evolution of this enzyme had to rely on random point mutations, it could have never evolved. Thus, if only 6 of these 47 mutations were essential for the evolution, the probability of achieving it in 30 years is about 3 x 10 -35. So, if the evolution could not be random, then it would have to be nonrandom, and as I have suggested in my book, they would be triggered by the environment. That is, the capability is built into the bacterium and the environment triggers the mutations.
I have ignored the evolution of enzyme 1, and the random evolution of that enzyme makes for an even less probable event.
Now, why should there be a built-in capability to metabolize nylon, which did not exist until 1937 or so? The answer is there shouldn't be. But there could have been a built-in capability to metabolize some other substrate. Kinoshita et al. (1981) tested enzyme 2 against 50 possible substrates and found no activity, but that does not mean that it doesn't have activity on some substrate not tested. The activity of enzyme 2 was small, but enabled the bacteria to metabolize the nylon waste. "
DISCUSSION
For starters, it appears that Spetner is suggesting that the mutations involved in the formation of nylonase enzymes were somehow caused by the environment, or made more likely because of the environment. But to make this case hold, Spetner should demonstrate that the 'right' mutations happened much more often than 'wrong' mutations. He's done nothing of the sort.
Additionally, Spetner's calculation of probability appears to assume that all the point mutations must occur simultaneously, which would give numbers like he specifies. However, he has produced no biological evidence that this supposed simultaneity is necessary.
Here are two key articles that counter Spetner's claims.
"Single-gene speciation by left–right reversal"
How many genetic mutations are required for the formation of a
new species? Is it 37? 140? 500?
This article, by Rei Ueshima and Takahiro Asami, appeared in Nature
425, 679, 16 October 2003, and shows that a mutation in ONE gene
in a snail was enough to produce sexual isolation, and thus a new
species. The gene controls the handedness of the snail's shell, and
right-handed forms simply cannot mate with left-handed ones. When
both right and left-handed specimens of the snail Euhadra were
found, DNA analyses were performed, and it was found that a
mutation in a single gene was responsible for the speciation
event.
(abstract,
PDF of
paper -
WARNING! GRAPHIC PICTURES OF SNAIL SEX)
"Functional proteins from a random-sequence library"
Spetner implies that the particular forms of nylonase studied must
have a precisely specified sequence for the entire enzyme. This
assumption is what drives his low-probability estimates, like the
figure of 3 x 10 -35 cited above. However, this paper by
Keefe and Szostak discusses a study of the likelihood of any
randomly-formed protein having significant biological activity. They
studied "a library of 6 x 1012 proteins each containing
80 contiguous random amino acids," and then "selected
functional proteins by enriching for those that bind to ATP."
Their conclusion: "...we suggest that functional proteins are
sufficiently common in protein sequence space (roughly 1 in
1011) that they may be discovered by entirely stochastic
means, such as presumably operated when proteins were first used by
living organisms. However, this frequency is still low enough to
emphasize the magnitude of the problem faced by those attempting de
novo protein design." In other words, the formation of functional
proteins is orders of magnitude more likely than Spetner's model
suggests.
PubMed
See Also: "A review of Lee Spetner's 'NOT BY CHANCE!'" by
Gert Korthof,
http://home.wxs.nl/~gkorthof/kortho36.htm
The author thanks Wesley Elsberry for useful discussions.
SPETNER has replied to these comments at Lodge's site, on October 27, 2004.
Spetner basically ignores the implications of the Keefe and Szostak paper discussed above, and claims that
"...the debate here is whether random mutations in the nylon bug generated the information that permitted it to metabolize the nylon waste or was there something nonrandom about it. By the latter I mean that either the correct mutations were induced by the environment or else the new adaptation was already built into the organism so that random mutations that would be likely to occur within the population could trigger the change.I am arguing that the above type of scenario could have occurred to account for the nylon-bug phenomenon, and he wants to argue that it could not have occurred that way and the only explanation is that random mutation can often lead to adaptation without any built-in preparation. Since these are the two sides of the argument, it seems unjust that he should require me to prove that the 'right' mutations (i.e. those induced by the environment) occur more often than those that don’t. On the contrary. He should have to prove that they do not occur more often. "
Spetner admits that "the mutation does yield an increase of information," but says this isn't a good example of evolution because the mutations involved may have been non-random, e.g. directed. So it all boils down to: Are Mutations Random?
It's time for Biology 101, folks - specifically, for the Evolution 101 course from UC Berkeley's dynamic "Understanding Evolution" website. If you follow the thread to Mechanisms, then Genetic Variation, and finally Mutations, you'll see a page titled "Mutations are Random." This page says, simply, that
Experimental data do not support directed mutation.If you expose bacteria to an antibiotic, you will likely observe an increased prevalence of antibiotic resistance. In 1952, Esther and Joshua Lederberg determined that these mutations do not happen because of exposure to the antibiotic. They showed that bacteria with a mutation for resistance existed in the population even before the population was exposed to the antibiotic—and that exposure to the antibiotic did not cause new resistant mutants to appear.
No known mechanism supports directed mutation.
Imagine what would have to happen in order for a mutation to be “directed” by the environment. Somehow, information about the chemicals in the lice shampoo would have to be conveyed to the DNA or cellular machinery. Then, something would have to select the appropriate changes to make in the DNA and make them. There are no known mechanisms that could do this—and it’s hard to even imagine a mechanism that could do this. Recall that science can only use natural explanations for natural phenomena.
The Berkeley site continues with an excellent lay discussion of The Lederberg Experiment, noting that
In 1952, Esther and Joshua Lederberg performed an experiment that helped to show that mutation is random, not directed.
I'm still digging around for examples of possible "directed" or "favored" mutations. If you know of some, send them in!
Read about more "Examples of Beneficial Mutations and Natural Selection" at
http://www.gate.net/~rwms/EvoMutations.html
![]()